Postdoctoral Research Associate
High Meadows Environmental Inst.
Princeton University

Photo: Jort Vanderveen

Dr. Nicole Nova is a Postdoctoral Research Associate at the High Meadows Environmental Institute at Princeton University. Dr. Nova received her PhD in Biology (Ecology and Evolution) from Stanford University in 2022. While at Stanford, she earned an MS in Statistics in 2020 and became a Stanford Data Science Scholar and a Predoctoral Fellow at the Center for Computational, Evolutionary and Human Genomics. Dr. Nova is a member of the Program for Disease Ecology, Health and the Environment and the Program for Conservation Genomics.
Dr. Nova combines mathematical modeling with statistics and data science to study the ecology and evolution of disease and conservation. She focuses on how climate change and other anthropogenic stressors impact health, biodiversity, and other aspects of the environment.
The goal of her research is to find environmentally sustainable win-win solutions for health and conservation. Dr. Nova aims to protect health, biodiversity, and conservation through research and policy.
Prior to her years at Stanford, Dr. Nova completed her undergraduate and graduate studies in dental surgery at Karolinska Institutet in Sweden. She also conducted biomedical research, and completed a Master's thesis on the pathophysiology of chronic pain in rheumatoid arthritis. This was conducted in the Molecular Pain Lab in collaboration with the Department of Physiology and Pharmacology and the Department of Oral Physiology at Karolinska Institutet.
She also studied abroad for a semester at Queen Mary University of London as an EU Erasmus Mundus scholar, and completed a clinical summer internship at the Department of Head and Neck Surgery, Medical University of Vienna, Austria. She worked on several head and neck cancer cases and became interested in the development of cancer at the cell population level. That spark of interest led her to study ecology and evolutionary biology, and its useful applications in medicine, public health, and eventually planetary health and conservation.
Dr. Nova decided to pursue a career in research after her first forays into academic research at Karolinska Institutet, Harvard and MIT as a high school student. Initially, she opted for combining research with clinical practice as a surgeon to help advancing medicine and improving health. In her early career as a surgeon, Dr. Nova became frustrated that she and other clinicians were often handling cases that were (or ultimately became) beyond repair. Most medical treatments were temporary and often had side effects, whereas her desire was to prevent many diseases at their origin. For example, the underlying cause of cancer or an emerging infectious disease is the spontaneous mutation of a single cell or pathogen, respectively. The progression of the disease is driven by complex evolutionary and ecological processes inside the body and outside in the environment. Thus, Dr. Nova decided to pursue PhD studies in ecology and evolutionary biology to seek sustainable solutions for disease prevention.
The study of evolutionary and ecological processes is highly quantitative. Throughout her life, Dr. Nova developed a strong affinity for mathematics and became fascinated by mathematical biology and its applications in medicine by modeling ecological and evolutionary phenomena. To strengthen her quantitative foundation, she completed coursework in mathematics and computer science at the Royal Institute of Technology in Sweden (here is a video of a robot that she and a team of engineering students built using Segway technology).
In 2014, Dr. Nova started working in the Michor Lab in the Department of Data Science at the Dana-Farber/Harvard Cancer Center (Dana-Farber Cancer Institute, Harvard Medical School, and Harvard T.H. Chan School of Public Health). She worked on mathematical modeling of cancer development using theories from evolutionary dynamics and population genetics.
In 2015, Dr. Nova started working as a Research Associate in the Koelle Research Group in the Department of Biology at Duke University. There she studied eco-evolutionary dynamics of infectious diseases. She formulated a mathematical model that captured the development of broadly neutralizing antibodies (BnAbs) in chronic HIV-infections. This allowed her to study the within-host co-evolution between BnAbs and HIV, which could potentially provide useful insights for developing effective HIV vaccines.
Over the years studying ecology and evolution, Dr. Nova became aware of the synergies between health and conservation. Climate change and human-modified landscapes often threaten both wildlife and human health. Raised next to a nature preserve in the Swedish woods, Dr. Nova gained a deep appreciation for protecting nature and the benefits that follow.



The overall research topic concerns conservation science and ecological/ evolutionary mechanisms for diseases in humans, domestic animals, and wildlife. Some of the ongoing research projects are described below. Other published research can be found under Publications.

This is a phylogenetic and phylodynamic study of a multi-host pathogen, called canine distemper virus. It has caused several outbreaks in the Yellowstone wolf population. It has also been reported in Arctic foxes in Alaska, but its disease dynamics in North American carnivores remains unknown. This study investigates the relatedness of canine distemper virus strains across different carnivore species in Yellowstone and Alaska to determine disease dynamics, transmission pathways, and potential reservoir host species. Because of its high mortality and rapid transmission, the virus poses a major threat to wild carnivores.
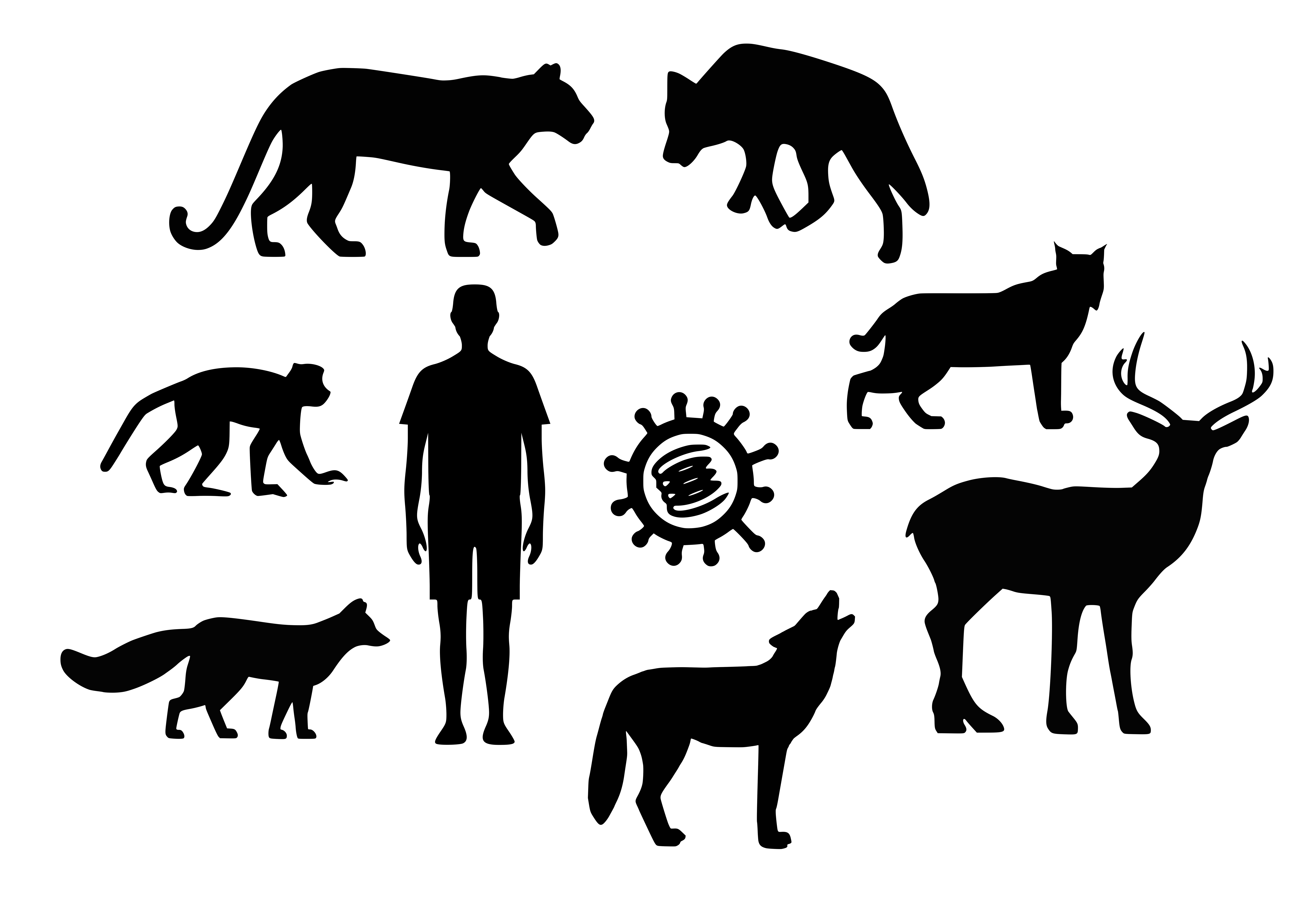
This is a project that determines when ecological factors (i.e., geography, host behavior, host life history, etc.) become more important for predicting parasite sharing between various host species than their phylogenetic relatedness (evolutionary factor). This is useful for predicting the most likely source of new emerging diseases threatening human health and the survival of endangered species.

* denotes co-first authorship
Holcomb KM, Mathis S, Staples JE, Fischer M, Barker CM, Beard CB, Nett RJ, Keyel AC, Marcantonio M, Childs ML, Gorris ME, Rochlin I, Hamins-Puertolas M, Ray EL, Uelman JA, DeFelice N, Freedman AS, Hollingsworth BD, Das P, Osthus D, Humphreys JM, Nova N, Mordecai EA, Cohnstaedt LW, Kirk DG, Kramer LD, Harris MJ, Kain MP, Reed EMX, Johansson MA. 2023.
Evaluation of an open forecasting challenge to assess skill of West Nile virus neuroinvasive disease prediction.
Parasites & Vectors. 16:11.
doi:10.1186/s13071-022-05630-y
Sokolow SH, Nova N, Jones IJ, Wood CL, Lafferty KD, Garchitorena A, Hopkins SR, Lund AJ, MacDonald AJ, Le Boa C, Peel AJ, Mordecai EA, Howard ME, Buck JC, Lopez-Carr D, Barry M, Bonds MH, De Leo GA. 2022. Ecological and socioeconomic factors associated with the human burden of environmentally mediated pathogens: a global analysis. The Lancet Planetary Health. 6(11):e870–e879.
doi:10.1016/S2542-5196(22)00248-0
Hopkins SR, Lafferty KD, Wood CL, Olson SH, Buck JC, De Leo GA, Fiorella KJ, Fornberg J, Garchitorena A, Jones IJ, Kuris AM, Kwong LH, LeBoa C, Leon AE, Lund AJ, MacDonald AJ, Metz DCG, Nova N, Peel AJ, Remais JV, Stewart Merrill TE, Wilson M, Bonds MH, Dobson AP, Lopez Carr D, Howard ME, Mandle L, Sokolow SH. 2022. Evidence gaps and diversity among potential win–win solutions for conservation and human infectious disease control. The Lancet Planetary Health. 6(8):e694–e705.
doi:10.1016/S2542-5196(22)00148-6
Hopkins SR*, Jones IJ*, Buck JC, LeBoa C, Kwong LH, Jacobsen K, Rickards C, Lund AJ, Nova N, MacDonald AJ, Lambert-Peck M, De Leo GA, Sokolow SH. 2022. Environmental persistence of the world’s most burdensome infectious and parasitic diseases. Frontiers in Public Health. 10:892366.
doi:10.3389/fpubh.2022.892366
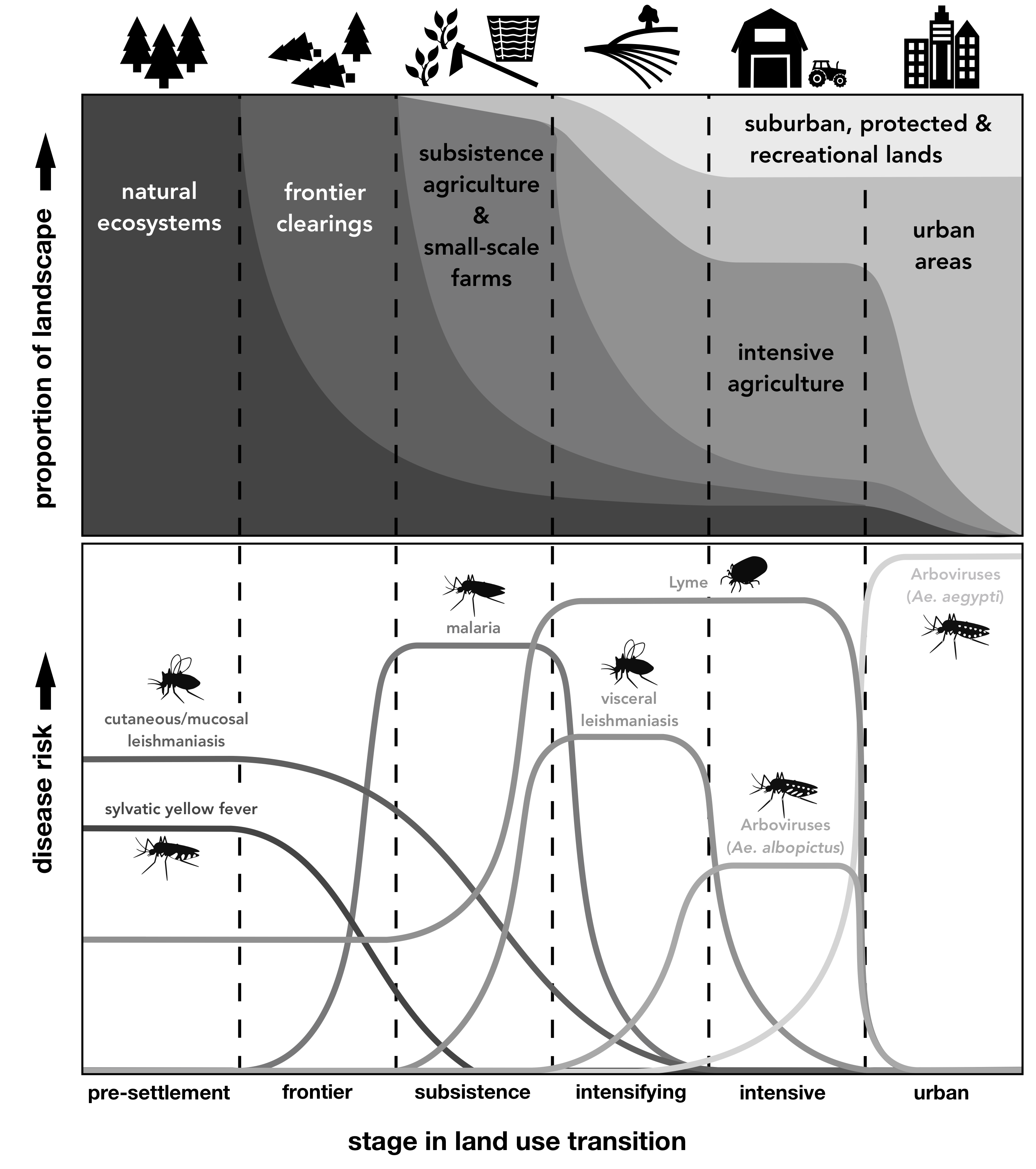
Nova N, Athni TS, Childs ML, Mandle L, Mordecai EA. 2022. Global change and emerging infectious diseases. Annual Review of Resource Economics. 14(1):333–354.
doi:10.1146/annurev-resource-111820-024214
Nova N, Pagliara R, Gordon DM. 2022. Individual variation does not regulate foraging response to humidity in harvester ant colonies. Frontiers in Ecology and Evolution. 9:756204. doi:10.3389/fevo.2021.756204
Glidden CK*, Nova N*, Kain MP, Lagerstrom KM, Skinner EB, Mandle L, Sokolow SH, Plowright RK, Dirzo R, De Leo GA, Mordecai EA. 2021.
Human-mediated impacts on biodiversity and the consequences for zoonotic disease spillover.
Current Biology. 31(19):R1342–R1361.
doi:10.1016/j.cub.2021.08.070
Nova N. 2021. Cross-species transmission of coronaviruses in humans and domestic mammals, what are the ecological mechanisms driving transmission, spillover, and disease emergence? Frontiers in Public Health. 9:717941.
doi:10.3389/fpubh.2021.717941
Childs ML*, Kain MP*, Harris MJ*, Kirk DG, Couper LI, Nova N, Delwel I, Ritchie J, Becker AD, Mordecai EA.
2021.
The impact of long-term non-pharmaceutical interventions on COVID-19 epidemic dynamics and control: the value and limitations of early models. Proceedings of the Royal Society B. 288(1957):20210811.
doi:10.1098/rspb.2021.0811
Couper LI, Farner JE, Caldwell JM, Childs ML, Harris MJ, Kirk DG, Nova N, Shocket MS, Skinner EB, Uricchio LH, Exposito-Alonso M, Mordecai EA.
2021.
How will mosquitoes adapt to climate warming?
eLife. 10:e69630.
doi:10.7554/eLife.69630
Hopkins SR, Sokolow SH, Buck JC, De Leo GA, Jones IJ, Kwong LH, LeBoa C, Lund AJ, MacDonald AJ, Nova N, Olson SH, Peel AJ, Wood CL, Lafferty KD. 2021. How to identify win–win interventions that benefit human health and conservation. Nature Sustainability. 4(4):298–304.
doi:10.1038/s41893-020-00640-z
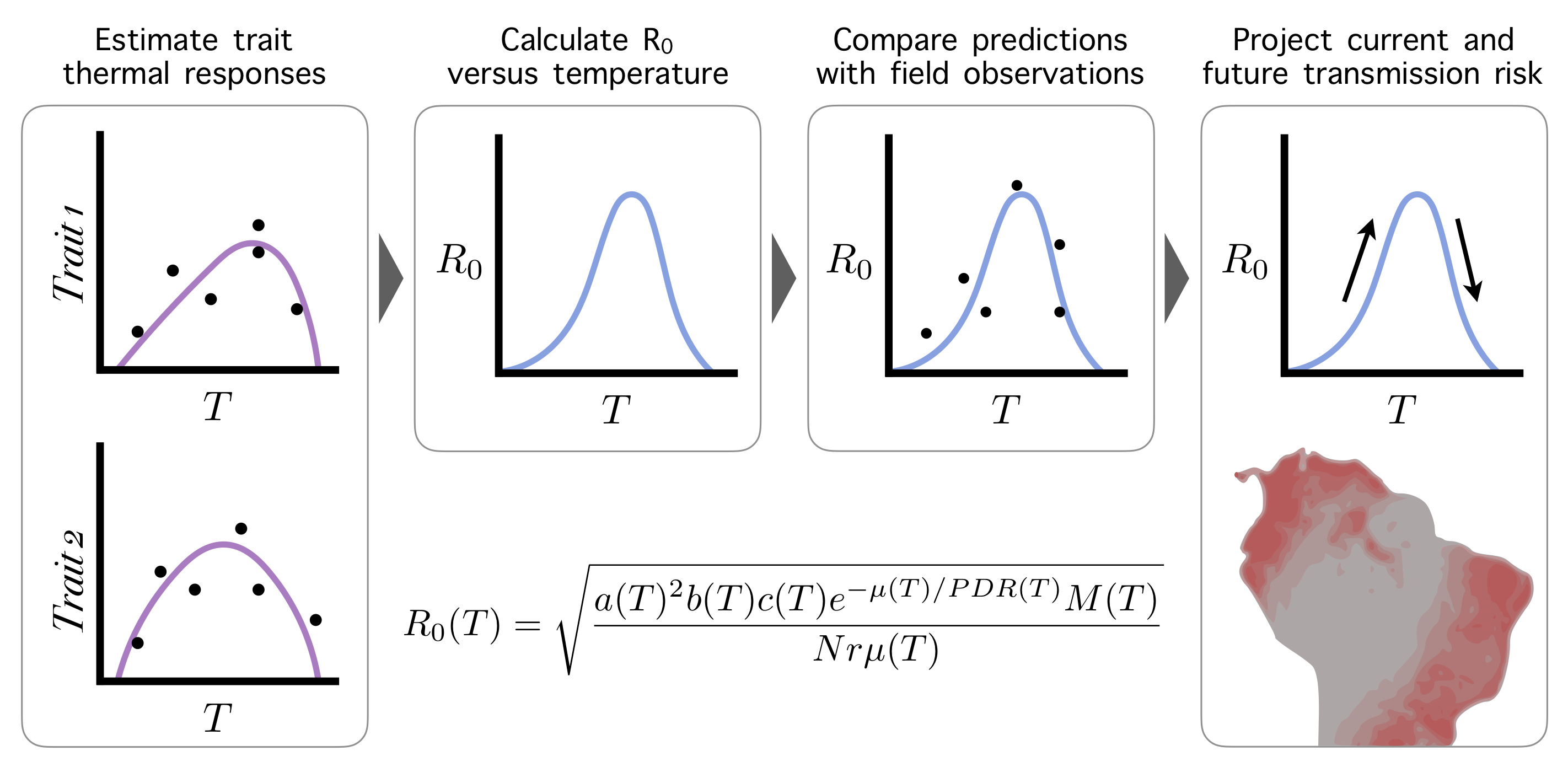
Athni TS, Shocket MS, Couper LI, Nova N, Caldwell IR, Caldwell JM, Childress JN, Childs ML, De Leo GA, Kirk D, MacDonald AJ, Olivarius K, Pickel DG, Winokur OC, Young HS, Cheng J, Grant EA, Kurzner PM, Kyaw S, Lin BJ, López RC, Massihpour DS, Olsen EC, Roache M, Ruiz A, Schultz EA, Shafat M, Spencer RL, Bharti N, Mordecai EA. 2021. The influence of vector-borne disease on human history: socio-ecological mechanisms. Ecology Letters. 24(4):829–846.
doi:10.1111/ele.13675
Nova N, Deyle ER, Shocket MS, MacDonald AJ, Childs ML, Rypdal M, Sugihara G, Mordecai EA. 2021. Susceptible host availability modulates climate effects on dengue dynamics. Ecology Letters. 24(3):415–425.
doi:10.1111/ele.13652
Allen WE*, Altae-Tran H*, Briggs J*, Jin X*, McGee G*, Shi A*, Raghavan R, Kamariza M,
Nova N, Pereta A, Danford C, Kamel A, Gothe P, Milam E, Aurambault J, Primke T, Li W, Inkenbrandt J, Huynh T, Chen E, Lee C, Croatto M, Bentley H, Lu W, Murray R, Travassos M, Coull BA, Openshaw J, Greene CS, Shalem O, King G, Probasco R, Cheng DR, Silbermann B, Zhang F, Lin X. 2020. Population-scale longitudinal mapping of COVID-19 symptoms, behaviour and testing. Nature Human Behaviour. 4(9):972–982.
doi:10.1038/s41562-020-00944-2
Smith JR, Hendershot JN, Nova N, Daily GC. 2020. The biogeography of ecoregions: Descriptive power across regions and taxa. Journal of Biogeography. 47(7):1413–1426. doi:10.1111/jbi.13871
Sokolow SH, Nova N, Pepin MK, Peel AJ, Pulliam JRC, Manlove K, Cross PC, Becker DJ, Plowright RK, McCallum H, De Leo GA. 2019. Ecological interventions to prevent and manage zoonotic pathogen spillover. Philosophical Transactions of the Royal Society B. 374(1782):20180342. doi:10.1098/rstb.2018.0342
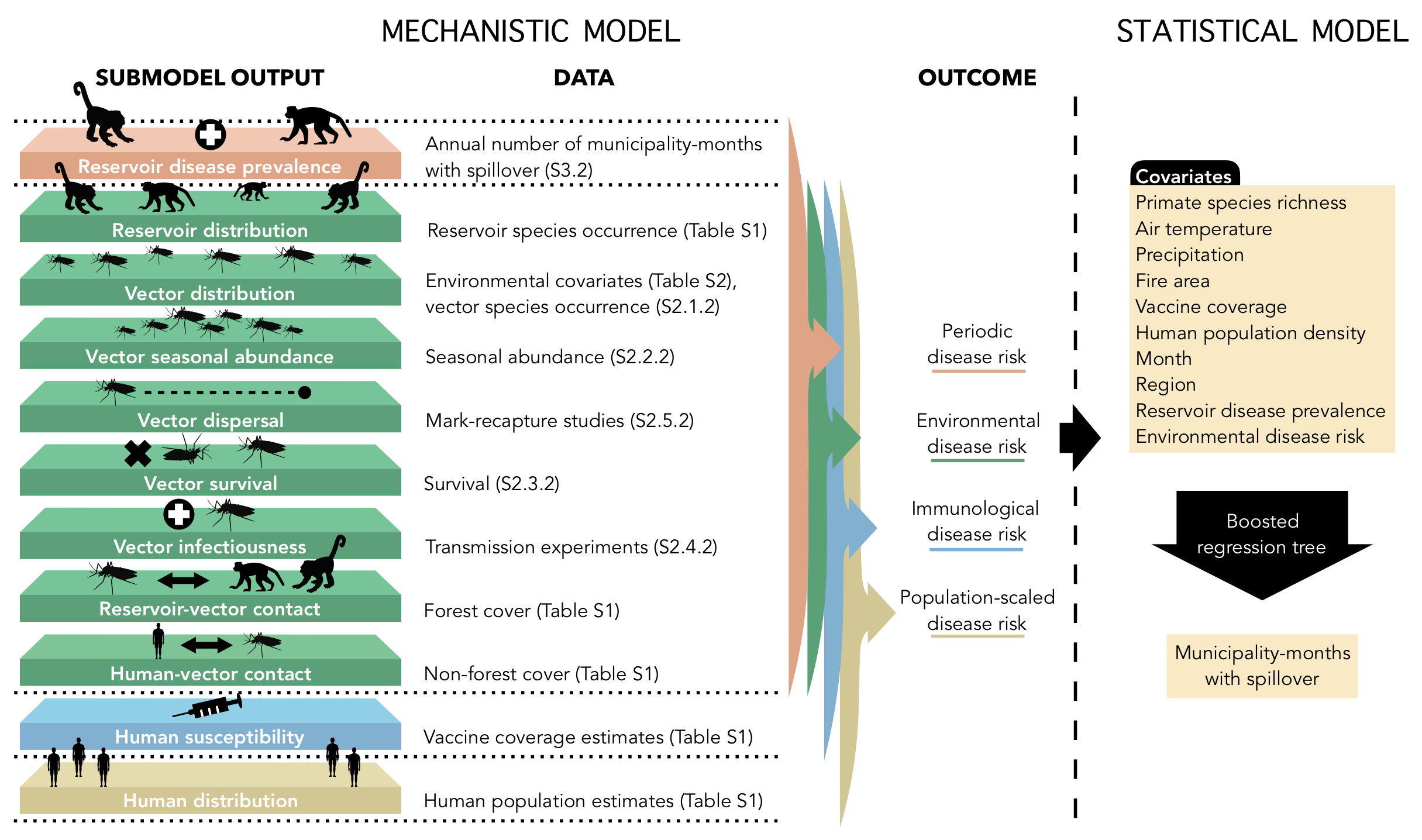
Childs ML, Nova N, Colvin J, Mordecai EA. 2019. Mosquito and primate ecology predict human risk of yellow fever virus spillover in Brazil. Philosophical Transactions of the Royal Society B. 374(1782):20180335. doi:10.1098/rstb.2018.0335
Leempoel K*, Nova N*, Meyer JM, Hebert T, Hadly EA. The natural return of an apex predator (Puma concolor) to a suburban preserve triggers a rapid behaviorally-mediated trophic cascade. In revision. bioRxiv preprint
Kirk DG*, Skinner EB*, Shocket MS, Couper LI, Nova N, Athni TS, Pourtois JD, Farner JE, Childs ML, Nyathi S, Mordecai EA. Climate Change and Disease Ecology. In: Suzán G, Aguirre AA, Mills JM, editors. The Ecology of Infectious Diseases: Methods on Evolution, Biodiversity, and Environmental Interactions. Oxford University Press. In press.
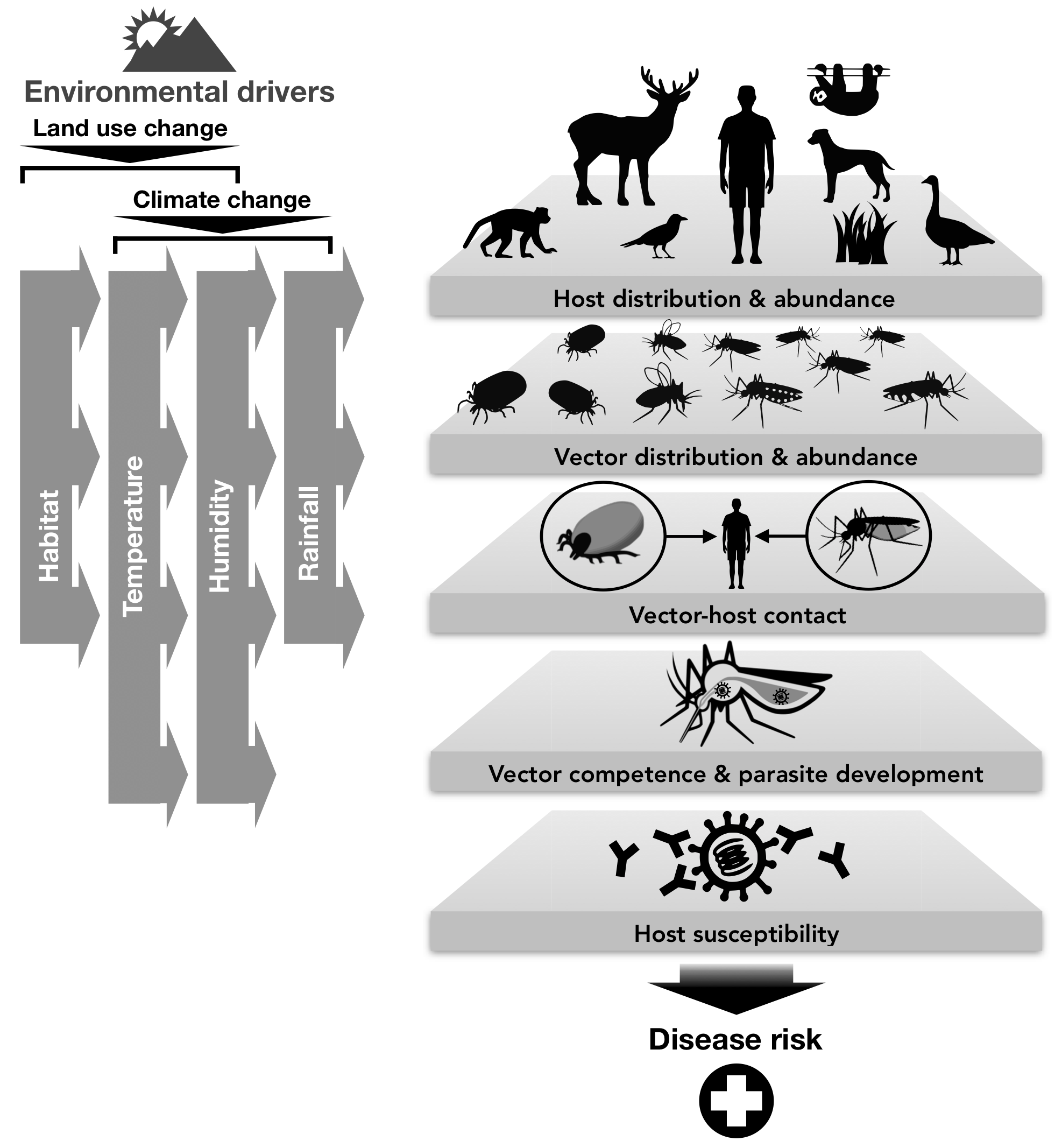
Shocket MS, Anderson CB, Caldwell JM, Childs ML, Couper LI, Han S, Harris MJ, Howard ME, Kain MP, MacDonald AJ, Nova N, Mordecai EA. 2021. Environmental drivers of vector-borne diseases. In: Drake JM, Bonsall M, Strand M, editors. Population Biology of Vector-borne Diseases (Ecology and Evolution of Infectious Diseases Series).
Oxford University Press.
ISBN:9780198853244
Van Wert M, Nova N, Horowitz T, Wolfe J. 2008. What does performance on one visual search task tell you about performance on another? Journal of Vision. 8(6):312. doi:10.1167/8.6.312
Nova N. 2022. Ecological and Evolutionary Mechanisms for Emerging Infectious Diseases: Case Studies in Humans and Mammalian Wildlife. PhD Dissertation, Department of Biology, Stanford University. Committee: Erin Mordecai, Dmitri Petrov, Elizabeth Hadly, Giulio De Leo, and Marcus Feldman.
PDF
Nova N. 2012. Chronic inflammation and pain: Assessment of c-Fos and ATF-3 as markers of spinal neuronal activity in a pain model of rheumatoid arthritis. MSc Thesis, Karolinska Institutet. Advisors: Per Alstergren and Camilla Svensson. PDF

Dr. Nova is also a graphic designer and scientific illustrator. Displayed here is an illustration named The Beauty of Mathematical Modeling in Biology that she created for a computer science class called Introduction to Computer Graphics and Imaging at Stanford University. Below is a sample portfolio of her artwork used for scientific publications or conferences.



RSI is a summer research program for high school students at Massachusetts Institute of Technology (MIT) and co-sponsored by the Center for Excellence in Education (CEE). Dr. Nova attended RSI as a student in 2007 and she worked with Prof. Jeremy Wolfe in the Visual Attention Lab in the Department of Brain and Cognitive Sciences at Harvard Medical School and Brigham and Women's Hospital. She was investigating whether a simple search task (i.e., looking for a green line among red lines) was correlated with the search for objects in the real world, in particular, the search for weapons in airport security (X-ray images of luggage). The work was later published in the Journal of Vision. As an RSI alum, Dr. Nova has been involved with the RSI program for several years and served as the Director of RSI in 2016, and a research mentor in 2020. For more information, see the CEE Alumni Spotlight press release page.
Rays is a summer research program for high school students, similar to Research Science Institute (RSI), but located in Sweden and it was founded in 2011. The first couple of years were run by Swedish RSI alumni (including Dr. Nova), before Rays alumni could also hold staff positions. Mentorships are being held at various universities in Stockholm, such as Karolinska Institutet, Stockholm University and the Royal Institute of Technology. After a summer-long internship in a research lab, the Rays students present their projects in the Swedish Museum of Science and Technology.